Survey Mask Software
- The survey masks
The survey masks distributed with the 2dFGRS Final Data Release are described here and in Colless et al. (2001) and Norberg et al. (2002). This version of the masks and mask software includes the mu-masks described in Colless et al. (2001) but not included in the original 100k release. It provides finer resolution in the masks, improvements to the speed of the code, and a correction for a minor bug that gave slightly incorrect positions for some of the holes in the survey.
- Copyright and use of the mask codes
As a user of the mask codes, we request that you observe the following guidelines:
- The code provided here has been written by several members of the 2dFGRS team. Peder Norberg and Shaun Cole (University of Durham, UK) wrote the software used to create the survey masks. If you use any part of this 2dFGRS software, please acknowledge "the 2dFGRS mask software by Peder Norberg and Shaun Cole".
- This code is supplied as-is and without guarantees - use it with caution and report any bugs.
- Read all of the documentation before trying to use the code!
- The mask data files
- Copy the following files:
- mask.ngp.dat (completeness mask for the NGP strip)
- mask.sgp.dat (completeness mask for the SGP strip)
- mask.ran.dat (completeness mask for the random fields)
- maglim.ngp.dat (magnitude limit mask for the NGP strip)
- maglim.sgp.dat (magnitude limit mask for the SGP strip)
- maglim.ran.dat (magnitude limit mask for the random fields)
- mumask.ngp.dat (mu-mask for the NGP strip)
- mumask.sgp.dat (mu-mask for the SGP strip)
- mumask.ran.dat (mu-mask for the random fields)
- Copy the following files:
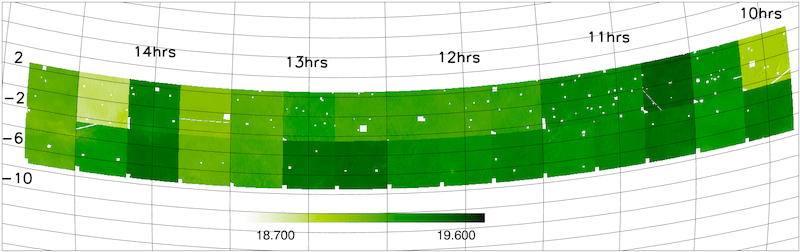
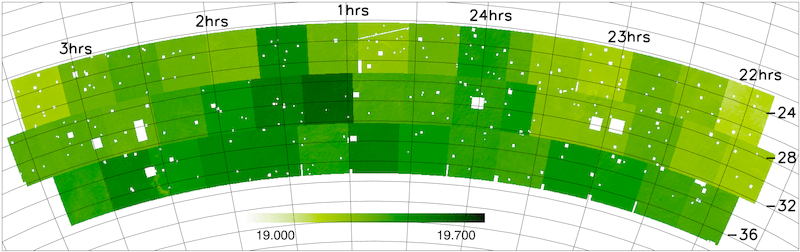
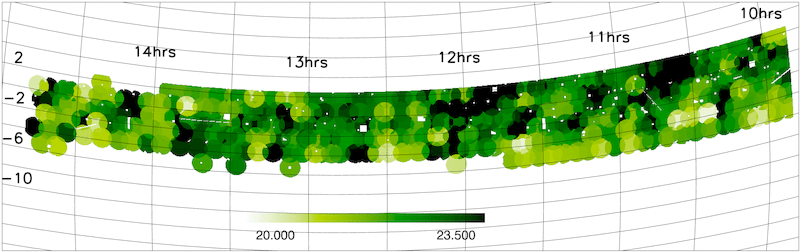
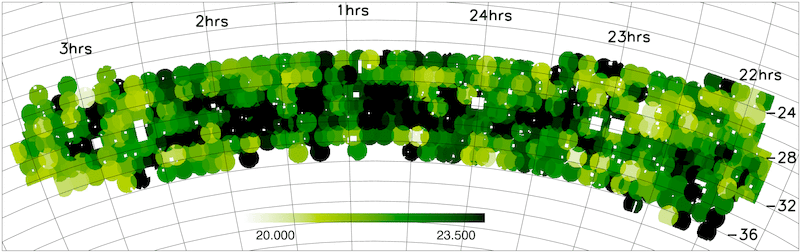
- Magnitude limit masks
The NGP and SGP magnitude limit masks are shown below.
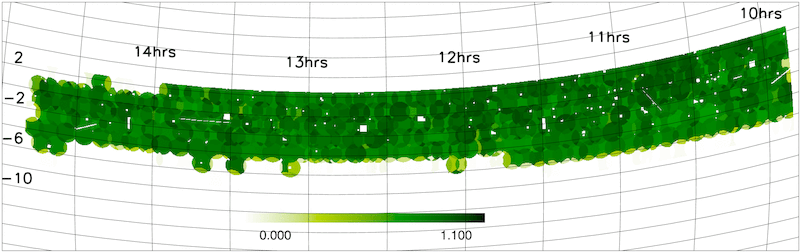
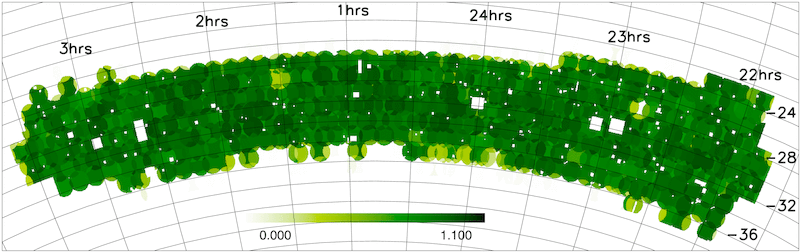
- Redshift completeness masks
The NGP and SGP redshift completeness masks are shown below.
- Mu-masks
The NGP and SGP mu-masks are shown below.
- Example code for using the masks
- Copy the following files:
- Choose the right compiler for your machine by selecting/deselecting the appropriate lines in the makefile. This code has been tested on Sun Solaris, DEC Alpha and PC Linux (with the Intel ifc compiler).
- Given R.A. and Dec. (B1950, in radians), release returns the completeness, the magnitude limit, the mu value and the UK Schmidt Telescope (UKST) sky survey plate number at that particular position. When compl = -1, the given position is outside the main 2dF boundary; when compl = -2, the given position is inside an excluded region (a 'hole').
-
Generating selected samples and mock catalogues
- Introduction: As well as the example code above, we also
provide code for selecting subsets of the survey data (using
various criteria defined by the user - see below) and generating
an associated unclustered catalogue of with the same set of
selection criteria. By changing various flags one can create
volume- or magnitude-limited catalogues, select galaxies within a
given range in absolute or apparent magnitude, choose only those
with a given spectral class or simply select galaxies within a
given redshift range or within regions of given completeness. The
main purpose of this code is the generation of the associated
distribution of unclustered galaxies with the same selection
criteria (a mock catalogue).
- Installing the code: Create a directory to work in and
copy to it
release_230k.tar.gz
(468Mb uncompressed, 50Mb compressed) and
aux_data.tar.gz
(168Mb uncompressed, 120Mb compressed). Gunzip and untar these
files. This will produce the following directories:
release_230k/ release_230k_sgp/ release_230k_ngp/ release_230k_ran/ aux_data/If you do not have Starlink libraries on your system, also copy to the working directory fitsio.tar.gz (1.5Mb uncompressed, 0.2Mb compressed) and sla.tar.gz (0.9Mb uncompressed, 0.2Mb compressed). The source code for these routines comes from the Starlink website, but has been slightly modifications so as to also work with the Intel ifc compiler under Linux. Gunzip and untar these to produce the following directories:fitsio/ sla/Finally, copy to the working directory select_release.tar.gz (0.8Mb uncompressed, 0.2Mb compressed). Gunzip and untar this to produce:readme_release.txt makefile select_release.com main/selection_release.f # sample selection and mock catalogue code sub/dist.for # subroutines used by the main codes sub/holes.f sub/in_2df_mask_select.f sub/indexx.f sub/iounix.f sub/locate.f sub/mag_error.f sub/pix_ran.inc sub/ran3.f sub/ran_all.f sub/ranmask.f sub/readascii.f sub/rtsafe.f sub/selfunc.f sub/tabulate_selfile.f sub/allheaders.dat # additional data files sub/distortion sub/ngpholes.lis sub/sgpholes.lis sub/rancentres.txt # data files for the random fields sub/ranfield_area.dat seldir/selfile.230k.2df_sgp # selection function files seldir/selfile.230k.2df_ngp seldir/selfile.230k.2df_ran seldir/selfile.230k.txt outdir/ # directory for outputChoose the right compiler for your machine by selecting/deselecting the appropriate lines in the makefile. This code has been tested on Sun, Alpha Dec and Linux (with the Intel ifc compiler).
- Running the code: The script
select_release.com
compiles the executable
select_release
and runs it for the parameters contained in the script. These
parameters, which can be changed by the user, are described (with
typical values) below:
# Switches with F (false) and T (true) options set dirp = release_230k # Directory from which to read the files set recalib = F # No influence for this version of code set mock = F # Selection made from a mock catalogue? set vlim = F # Create a volume limited sample? set mag_fix = F # Use fixed magnitude limit? set mu_mask = T # Use mu mask correction? set magerr = F # Apply magnitude errors? set zzerr = F # Apply redshift errors? set spec = F # Using spectral class information? # Basic selection criteria set zmin = 0.002 # Minimum redshift considered set zmax = 0.3 # Maximum redshift considered set magmin = 15.0 # Bright apparent magnitude considered set magmax = 19.35 # Faint apparent magnitude considered # (has no effect unless mag_fix = T) set compl_cut = 0.5 # Sector completeness cut set iseed = -75843 # Initial random seed (needs to be # changed when creating a set of # different mock catalogues) set ng_ranmult = 1 # Multiple of the number of galaxies # produced for the mock catalogues # (if <= 0, no mocks are produced) set region = sgp # Region analyzed: ngp, sgp or ran # The value of region is correctly assigned (within the code) to # pick up the appropriate input files # Names of various mask files that are read in set maskfile = $dirp"_reg/mask.dat" set maglimmask = $dirp"/maglim_mask.reg.dat" set mumaskfile = $dirp"_reg/mask_mu.dat" set selfile = seldir/selfile.$dir.2df_reg # Selection function file set catname = $dirp"/parent.reg.txt" # 2dFGRS catalogue set file1 = $dirp"_reg/weight.txt" # 1st list of weights set file2 = $dirp"_reg/weight_extra.txt" # 2nd list of weights # Names of output files: set outfile1 = "outdir/select_real_"$region".out" # selected data set outfile2 = "outdir/select_mock_"$region".out" # mock catalogue # Additional selection criteria useful only if you want to # select volume limited subsets of the data, possibly with # additional selection on magnitudes and eta: set nlim = 0 # nlim=0 : values of lim_spec (below) ignored # nlim=1 : construct a volume limited catalogue defined by # the absolute magnitude limits lim_spec1 & lim_spec2 # nlim=2 : within the volume limit select only galaxies with # absolute magnitudes between lim_spec3 and lim_spec4 # nlim=3 : within this subset select only galaxies with # eta value between lim_spec5 and lim_spec6 # This pair of parameters define the limit of the volume set lim_spec1 = -20.5 # Bright absolute magnitude limit set lim_spec2 = -20.0 # Faint absolute magnitude limit # This pair define the absolute magnitude range of the galaxy # sample that is returned. These magnitude limits must lie within # the magnitude range (lim_spec1,lim_spec2) that defines the # sample volume. set lim_spec3 = -20.5 # Bright absolute magnitude limit set lim_spec4 = -20.0 # Faint absolute magnitude limit # This pair allow selection using spectral class information set lim_spec5 = -5.0 # Lower eta limit set lim_spec6 = +60.0 # Upper eta limit - Inputs: The main input is the galaxy input catalogue
($dirp/parent.reg.txt).
The format of this file is explained in the
description
of the spectral catalogues. Other versions can be created fairly
straightforwardly from the
mSQL database.
The other input files will always be the sam ( provided you don't
change the pixel scale) and therefore do not need to be updated.
Currently the pixel scale is set to 1.4arcmin x 1.4arcmin, but can
be improved by changing the variables
NPX and
NPY (defined in the
main programs) accordingly; 1.4' x 1.4' corresponds to
NPX=2*1800
and NPY=2*600.
Note that the pixel scale needs to be the same as the one used in
generating the masks.
- Outputs:
select_release.exe
outputs two files - one for the selected data (in
outfile1) and one
for the randoms (in
outfile2). If you
want to change the format of these two files, you can do so by
setting the code variable
write_cat=.false.
and inserting your own code at the bottom of
program selectdata
as indicated in the code. The standard format of the output is:
ra = galaxy R.A. (B1950) dec = galaxy Dec. (B1950) zz = best galaxy redshift mag = galaxy b_J magnitude wsel = weight assigned by 10 nearest neighbours (equal to 1 if all neighbours have a good quality redshift; typically equal to 2 if all 10 nearest neighbours are missing) compl = completeness of associated sector selz = selection function value for galaxy (without mu_mask) selz_mu = selection function value for galaxy (with mu_mask) bjlim = b_J magnitude limit serial = galaxy serial number (only for real data, not mocks; identical to the one listed in $dirp/parent.reg.txt) - Notes on the code:
- Some additional files are produced by these programs and used at different stages of the analysis; these files are not relevant to the user.
- There are a few parameters that are set within the main program rather than in the script. These have been set to typical values, but can be changed at the user's discretion (and risk).
- The program uses a Omega=0.3, Lambda=0.7 cosmology by default.
- Introduction: As well as the example code above, we also
provide code for selecting subsets of the survey data (using
various criteria defined by the user - see below) and generating
an associated unclustered catalogue of with the same set of
selection criteria. By changing various flags one can create
volume- or magnitude-limited catalogues, select galaxies within a
given range in absolute or apparent magnitude, choose only those
with a given spectral class or simply select galaxies within a
given redshift range or within regions of given completeness. The
main purpose of this code is the generation of the associated
distribution of unclustered galaxies with the same selection
criteria (a mock catalogue).
- Limitations and corrections
- Each pixel in the masks is 1.4 arcmin on a side. This is much
larger than the precision of the 2dFGRS galaxy positions. Hence
the masks can indicate that some 2dFGRS galaxies are outside the
survey boundary or inside excluded holes due to pixelisation
effects, as the masks have been defined in a conservative
way.
- In the definition of the magnitude-dependent completeness mask in
section 8.3 of Colless et al. (2001), there is an error in the
value given for the parameter alpha; the paper gives alpha=0.5,
but the correct value is alpha=0.5ln(10).
Please report any serious problems encountered with this software to the 2dFGRS team.
- Each pixel in the masks is 1.4 arcmin on a side. This is much
larger than the precision of the 2dFGRS galaxy positions. Hence
the masks can indicate that some 2dFGRS galaxies are outside the
survey boundary or inside excluded holes due to pixelisation
effects, as the masks have been defined in a conservative
way.